publications
(*) denotes equal contribution
2025
- MIDL
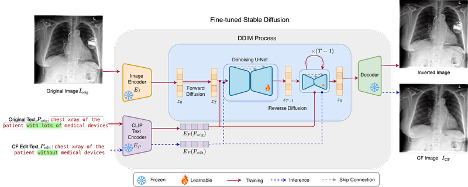 Prism: High-resolution & precise counterfactual medical image generation using language-guided stable diffusionAmar Kumar, Anita Kriz, Mohammad Havaei, and 1 more authorMIDL, 2025
Prism: High-resolution & precise counterfactual medical image generation using language-guided stable diffusionAmar Kumar, Anita Kriz, Mohammad Havaei, and 1 more authorMIDL, 2025Runner-up - Best Oral Paper
Developing reliable and generalizable deep learning systems for medical imaging faces significant obstacles due to spurious correlations, data imbalances, and limited text annotations in datasets. Addressing these challenges requires architectures that are robust to the unique complexities posed by medical imaging data. Rapid advancements in visionlanguage foundation models within the natural image domain prompt the question of how they can be adapted for medical imaging tasks. In this work, we present PRISM, a framework that leverages foundation models to generate high-resolution, language-guided medical image counterfactuals using Stable Diffusion. Our approach demonstrates unprecedented precision in selectively modifying spurious correlations (the medical devices) and disease features, enabling the removal and addition of specific attributes while preserving other image characteristics. Through extensive evaluation, we show how PRISM advances counterfactual generation and enables the development of more robust downstream classifiers for clinically deployable solutions. To facilitate broader adoption and research, we make our code publicly available at https://github.com/Amarkr1/PRISM.
- MICCAI
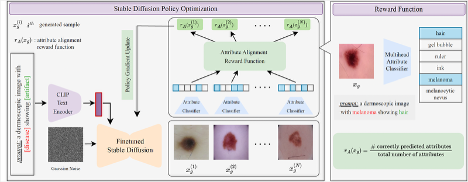 Rl4med-ddpo: Reinforcement learning for controlled guidance towards diverse medical image generation using vision-language foundation modelsParham Saremi*, Amar Kumar*, Mohamed Mohamed, and 2 more authorsMICCAI, 2025
Rl4med-ddpo: Reinforcement learning for controlled guidance towards diverse medical image generation using vision-language foundation modelsParham Saremi*, Amar Kumar*, Mohamed Mohamed, and 2 more authorsMICCAI, 2025Vision-Language Foundation Models (VLFM) have shown a tremendous increase in performance in terms of generating high-resolution, photorealistic natural images. While VLFMs show a rich understanding of semantic content across modalities, they often struggle with fine-grained alignment tasks that require precise correspondence between image regions and textual descriptions, a limitation in medical imaging, where accurate localization and detection of clinical features are essential for diagnosis and analysis. To address this issue, we propose a multi-stage architecture where a pre-trained VLFM (e.g. Stable Diffusion) provides a cursory semantic understanding, while a reinforcement learning (RL) algorithm refines the alignment through an iterative process that optimizes for understanding semantic context. The reward signal is designed to align the semantic information of the text with synthesized images. Experiments on the public ISIC2019 skin lesion dataset demonstrate that the proposed method improves (a) the quality of the generated images, and (b) the alignment with the text prompt over the original fine-tuned Stable Diffusion baseline. We also show that the synthesized samples could be used to improve disease classifier performance for underrepresented subgroups through augmentation. Our code is accessible through the project website https://parhamsaremi.github.io/rl4med-ddpo
- MICCAIAURA: A Multi-Modal Medical Agent for Understanding, Reasoning & AnnotationNima Fathi, Amar Kumar, and Tal ArbelarXiv preprint arXiv:2507.16940, 2025
Recent advancements in Large Language Models (LLMs) have catalyzed a paradigm shift from static prediction systems to agentic AI—intelligent agents capable of reasoning, interacting with tools, and adapting to complex tasks. While LLM agentic systems have shown promise across many domains, their application to medical imaging remains in its infancy. In this work, we introduce AURA, the first visual linguistic explainability agent designed specifically for comprehensive analysis, explanation, and evaluation of medical images. By enabling dynamic interactions, contextual explanations, and hypothesis testing, AURA represents a significant advancement toward more transparent, adaptable, and clinically aligned AI systems. This work highlights the promise of agentic AI in transforming medical image analysis from static predictions to interactive decision support. Leveraging Qwen-32B, an LLM-based architecture, AURA integrates a modular toolbox comprising: (i) a segmentation suite with phase grounding, pathology segmentation, and anatomy segmentation to localize clinically meaningful regions; (ii) a counterfactual image generation module that supports reasoning through image-level explanations; and (iii) a set of evaluation tools, including pixel-wise difference map analysis, classification, and advanced state-of-the-art components, to assess the diagnostic relevance and visual interpretability of the results.
- MIDLPixel Perfect MegaMed: A Megapixel-Scale Vision-Language Foundation Model for Generating High Resolution Medical ImagesZahra TehraniNasab, Amar Kumar, and Tal ArbelarXiv preprint arXiv:2507.12698, 2025
Medical image synthesis presents unique challenges due to the inherent complexity and high-resolution details required in clinical contexts. Traditional generative architectures such as Generative Adversarial Networks (GANs) or Variational Auto Encoder (VAEs) have shown great promise for high-resolution image generation but struggle with preserving fine-grained details that are key for accurate diagnosis. To address this issue, we introduce Pixel Perfect MegaMed, the first vision-language foundation model to synthesize images at resolutions of 1024x1024. Our method deploys a multi-scale transformer architecture designed specifically for ultra-high resolution medical image generation, enabling the preservation of both global anatomical context and local image-level details. By leveraging vision-language alignment techniques tailored to medical terminology and imaging modalities, Pixel Perfect MegaMed bridges the gap between textual descriptions and visual representations at unprecedented resolution levels. We apply our model to the CheXpert dataset and demonstrate its ability to generate clinically faithful chest X-rays from text prompts. Beyond visual quality, these high-resolution synthetic images prove valuable for downstream tasks such as classification, showing measurable performance gains when used for data augmentation, particularly in low-data regimes. Our code is accessible through the project website - https://tehraninasab.github.io/pixelperfect-megamed/.
- CVPRLeveraging Vision-Language Foundation Models to Reveal Hidden Image-Attribute Relationships in Medical ImagingAmar Kumar, Anita Kriz, Barak Pertzov, and 1 more authorIn Workshop Proceedings of the Computer Vision and Pattern Recognition Conference, 2025
Vision-language foundation models (VLMs) have shown impressive performance in guiding image generation through text, with emerging applications in medical imaging. In this work, we are the first to investigate the question: ’Can fine-tuned foundation models help identify critical, and possibly unknown, data properties?’ By evaluating our proposed method on a chest x-ray dataset, we show that these models can generate high-resolution, precisely edited images compared to methods that rely on Structural Causal Models (SCMs) according to numerous metrics. For the first time, we demonstrate that fine-tuned VLMs can reveal hidden data relationships that were previously obscured due to available metadata granularity and model capacity limitations. Our experiments demonstrate both the potential of these models to reveal underlying dataset properties while also exposing the limitations of fine-tuned VLMs for accurate image editing and susceptibility to biases and spurious correlations.
- CVPRLanguage-Guided Trajectory Traversal in Disentangled Stable Diffusion Latent Space for Factorized Medical Image GenerationZahra TehraniNasab, Amar Kumar, and Tal ArbelIn Workshop Proceedings of the Computer Vision and Pattern Recognition Conference, 2025
Text-to-image diffusion models have demonstrated a remarkable ability to generate photorealistic images from natural language prompts. These high-resolution, language-guided synthesized images are essential for the explainability of disease or exploring causal relationships. However, their potential for disentangling and controlling latent factors of variation in specialized domains like medical imaging remains under-explored. In this work, we present the first investigation of the power of pre-trained vision-language foundation models, once fine-tuned on medical image datasets, to perform latent disentanglement for factorized medical image generation and interpolation. Through extensive experiments on chest X-ray and skin datasets, we illustrate that fine-tuned, language-guided Stable Diffusion inherently learns to factorize key attributes for image generation, such as the patient’s anatomical structures or disease diagnostic features. We devise a framework to identify, isolate, and manipulate key attributes through latent space trajectory traversal of generative models, facilitating precise control over medical image synthesis.
2024
- MIDLDecodex: Confounder detector guidance for improved diffusion-based counterfactual explanationsNima Fathi, Amar Kumar, Brennan Nichyporuk, and 2 more authorsarXiv preprint arXiv:2405.09288, 2024
Deep learning classifiers are prone to latching onto dominant confounders present in a dataset rather than on the causal markers associated with the target class, leading to poor generalization and biased predictions. Although explainability via counterfactual image generation has been successful at exposing the problem, bias mitigation strategies that permit accurate explainability in the presence of dominant and diverse artifacts remain unsolved. In this work, we propose the DeCoDEx framework and show how an external, pre-trained binary artifact detector can be leveraged during inference to guide a diffusion-based counterfactual image generator towards accurate explainability. Experiments on the CheXpert dataset, using both synthetic artifacts and real visual artifacts (support devices), show that the proposed method successfully synthesizes the counterfactual images that change the causal pathology markers associated with Pleural Effusion while preserving or ignoring the visual artifacts. Augmentation of ERM and Group-DRO classifiers with the DeCoDEx generated images substantially improves the results across underrepresented groups that are out of distribution for each class. The code is made publicly available at https://github.com/NimaFathi/DeCoDEx.
2023
- MICCAIDebiasing counterfactuals in the presence of spurious correlationsAmar Kumar, Nima Fathi, Raghav Mehta, and 4 more authorsIn Workshop on Clinical Image-Based Procedures, 2023
Deep learning models can perform well in complex medical imaging classification tasks, even when basing their conclusions on spurious correlations (i.e. confounders), should they be prevalent in the training dataset, rather than on the causal image markers of interest. This would thereby limit their ability to generalize across the population. Explainability based on counterfactual image generation can be used to expose the confounders but does not provide a strategy to mitigate the bias. In this work, we introduce the first end-to-end training framework that integrates both (i) popular debiasing classifiers (e.g. distributionally robust optimization (DRO)) to avoid latching onto the spurious correlations and (ii) counterfactual image generation to unveil generalizable imaging markers of relevance to the task. Additionally, we propose a novel metric, Spurious Correlation Latching Score (SCLS), to quantify the extent of the classifier reliance on the spurious correlation as exposed by the counterfactual images. Through comprehensive experiments on two public datasets (with the simulated and real visual artifacts), we demonstrate that the debiasing method: (i) learns generalizable markers across the population, and (ii) successfully ignores spurious correlations and focuses on the underlying disease pathology.
2022
- MICCAICounterfactual image synthesis for discovery of personalized predictive image markersAmar Kumar, Anjun Hu, Brennan Nichyporuk, and 4 more authorsIn MICCAI Workshop on Medical Image Assisted Blomarkers’ Discovery, 2022
The discovery of patient-specific imaging markers that are predictive of future disease outcomes can help us better understand individual-level heterogeneity of disease evolution. In fact, deep learning models that can provide data-driven personalized markers are much more likely to be adopted in medical practice. In this work, we demonstrate that data-driven biomarker discovery can be achieved through a counterfactual synthesis process. We show how a deep conditional generative model can be used to perturb local imaging features in baseline images that are pertinent to subject-specific future disease evolution and result in a counterfactual image that is expected to have a different future outcome. Candidate biomarkers, therefore, result from examining the set of features that are perturbed in this process. Through several experiments on a large-scale, multi-scanner, multi-center multiple sclerosis (MS) clinical trial magnetic resonance imaging (MRI) dataset of relapsing-remitting (RRMS) patients, we demonstrate that our model produces counterfactuals with changes in imaging features that reflect established clinical markers predictive of future MRI lesional activity at the population level. Additional qualitative results illustrate that our model has the potential to discover novel and subject-specific predictive markers of future activity.